DAVID Ortholog
Introduction
DAVID Ortholog is used for the conversion of gene lists between species using the ortholog pair data from Orthologous MAtrix (OMA) and Ensembl Compara which has been integrated into the DAVID Knowledgebase. DAVID Ortholog can convert a gene list to an ortholog list of a targetted species and the downstream DAVID analysis, in the context of that targetted species, may continue seamlessly using any available DAVID analyses tools. This can make it easy for users to further understand the biological meaning of their gene list based on the functional annotation of the orthologs.
Step 2
After uploading the study gene/protein list and selecting the DAVID Ortholog tool, the Knowledgebase is queried to determine which taxonomies have orthologs for genes in the study list. After being presented with this list of potential target species, the user will select one target species to continue the conversion. A search feature is available to aid users in selecting the ortholog species.

Step 3
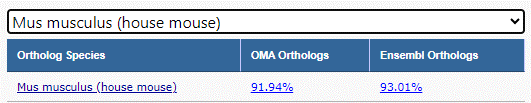
After the user selects the ortholog taxonomy of interest, the Knowledgebase is again queried in real time to determine the percentage of genes in the list with orthologs from OMA and Ensembl. The user will now select the ortholg data source (OMA/Ensembl) with which they would like to continue the conversion.
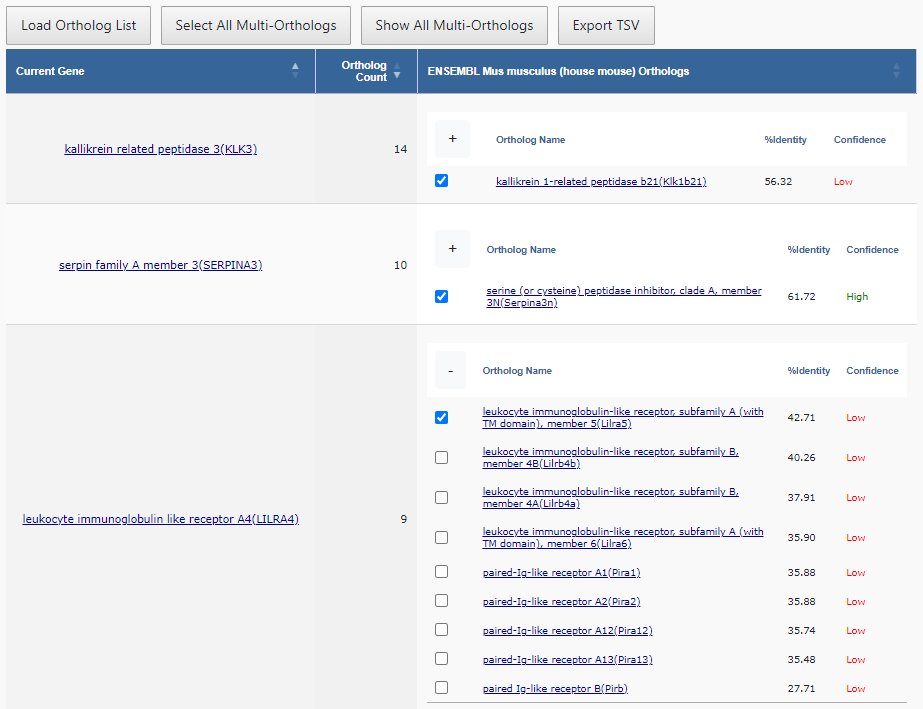
Step 4
Once the user selects the ortholog data source, the Knowledgebase is again queried to retrieve the orthologs for the target ortholog taxonomy and data source. The user may now choose which genes to convert and load into DAVID for further analyses in the context of the target ortholog taxonomy. The download mechanism is called after the data view and the download button appears when results are ready.
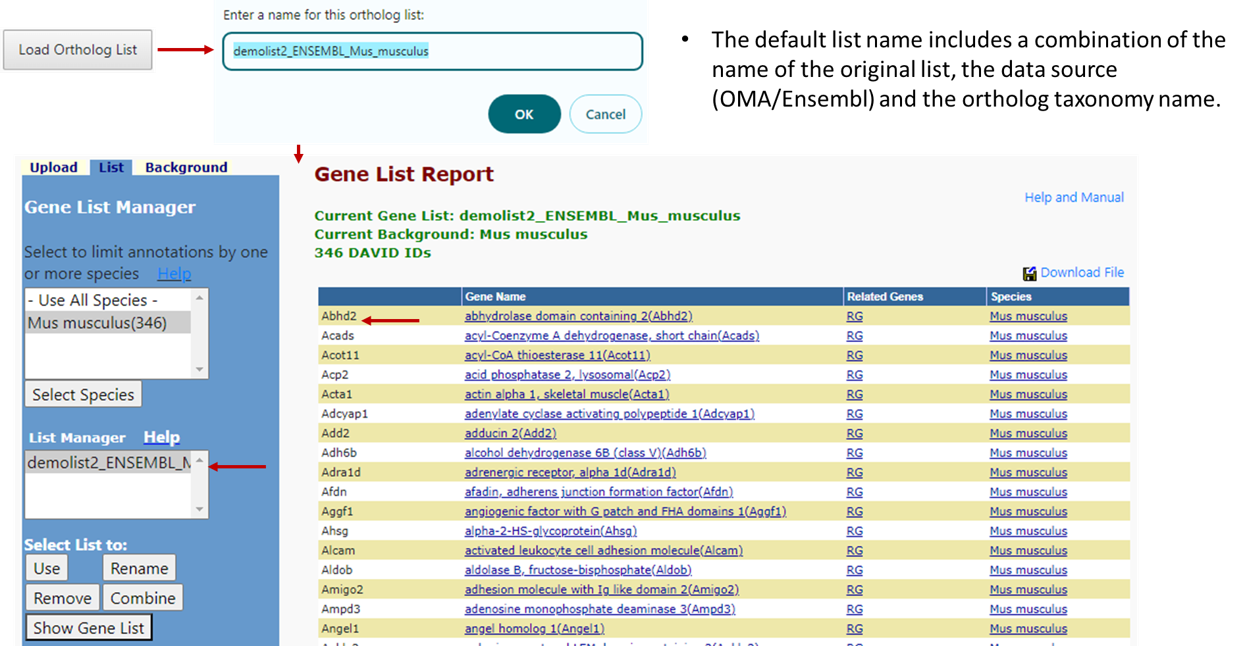
Step 5
Once all selections are made, the user will click the "Load Ortholog List" button and will be prompted to name the new ortholog list which by default is a combination of the name of the original list, the data source (OMA/Ensembl) and the ortholog taxonomy name. After the list name is submitted, the orthologs will be loaded into the user's workspace as a new and current list along with all ortholog species genes being loaded as the background population for further analyses with any available tools in DAVID.